I have the following data:
simres_auc2 <- structure(list(MINDGDP = c(1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L, 1L,
1L, 1L, 1L, 1L, 1L, 1L, 1L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L, 2L,
2L, 2L, 2L, 2L, 2L, 2L, 2L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L, 3L,
3L, 3L, 3L, 3L, 3L, 3L, 3L), PSIZE = c(5L, 5L, 5L, 5L, 10L, 10L,
10L, 10L, 20L, 20L, 20L, 20L, 50L, 50L, 50L, 50L, 5L, 5L, 5L,
5L, 10L, 10L, 10L, 10L, 20L, 20L, 20L, 20L, 50L, 50L, 50L, 50L,
5L, 5L, 5L, 5L, 10L, 10L, 10L, 10L, 20L, 20L, 20L, 20L, 50L,
50L, 50L, 50L), simno = c(13L, 13L, 13L, 13L, 16L, 16L, 16L,
16L, 19L, 19L, 19L, 19L, 22L, 22L, 22L, 22L, 13L, 13L, 13L, 13L,
16L, 16L, 16L, 16L, 19L, 19L, 19L, 19L, 22L, 22L, 22L, 22L, 13L,
13L, 13L, 13L, 16L, 16L, 16L, 16L, 19L, 19L, 19L, 19L, 22L, 22L,
22L, 22L), METHOD_RED = c("EVA (alpha = 0.001)", "EVA (alpha = 0.005)",
"EVA (alpha = 0.01)", "EVA (alpha = 0.05)", "EVA (alpha = 0.001)",
"EVA (alpha = 0.005)", "EVA (alpha = 0.01)", "EVA (alpha = 0.05)",
"EVA (alpha = 0.001)", "EVA (alpha = 0.005)", "EVA (alpha = 0.01)",
"EVA (alpha = 0.05)", "EVA (alpha = 0.001)", "EVA (alpha = 0.005)",
"EVA (alpha = 0.01)", "EVA (alpha = 0.05)", "EVA (alpha = 0.001)",
"EVA (alpha = 0.005)", "EVA (alpha = 0.01)", "EVA (alpha = 0.05)",
"EVA (alpha = 0.001)", "EVA (alpha = 0.005)", "EVA (alpha = 0.01)",
"EVA (alpha = 0.05)", "EVA (alpha = 0.001)", "EVA (alpha = 0.005)",
"EVA (alpha = 0.01)", "EVA (alpha = 0.05)", "EVA (alpha = 0.001)",
"EVA (alpha = 0.005)", "EVA (alpha = 0.01)", "EVA (alpha = 0.05)",
"EVA (alpha = 0.001)", "EVA (alpha = 0.005)", "EVA (alpha = 0.01)",
"EVA (alpha = 0.05)", "EVA (alpha = 0.001)", "EVA (alpha = 0.005)",
"EVA (alpha = 0.01)", "EVA (alpha = 0.05)", "EVA (alpha = 0.001)",
"EVA (alpha = 0.005)", "EVA (alpha = 0.01)", "EVA (alpha = 0.05)",
"EVA (alpha = 0.001)", "EVA (alpha = 0.005)", "EVA (alpha = 0.01)",
"EVA (alpha = 0.05)"), auc = c(0.5, 0.440423333333333, 0.73412,
0.570526, 0.5, 0.465404, 0.695695333333333, 0.536143333333333,
0.5, 0.482674, 0.673217333333333, 0.517231333333333, 0.5, 0.478126666666667,
0.661129333333333, 0.530846, 0.5, 0.4520975, 0.742583, 0.577082,
0.5, 0.4546035, 0.694907, 0.550087, 0.5, 0.4706495, 0.6585825,
0.544709, 0.5, 0.473219, 0.659395, 0.546985, 0.5, 0.45364, 0.754459333333333,
0.58385, 0.5, 0.442713333333333, 0.699316, 0.563635333333333,
0.5, 0.486780666666667, 0.678044666666667, 0.554051333333333,
0.5, 0.462297333333333, 0.651185333333333, 0.544234666666667)), class = c("tbl_df",
"tbl", "data.frame"), row.names = c(NA, -48L), .Names = c("MINDGDP",
"PSIZE", "simno", "METHOD_RED", "auc"))
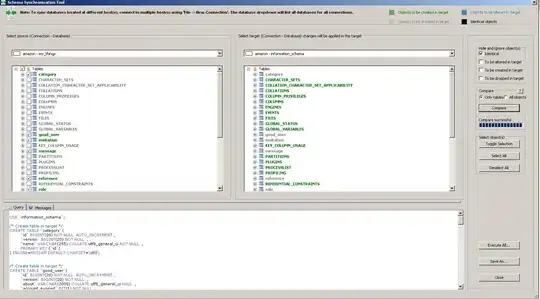
The following code generates the following plot, where position_dodge is working correctly.
ggplot2::ggplot(data = simres_auc2,
aes_string(x = "factor(METHOD_RED)",
y = "auc")) +
ggplot2::geom_point(aes_string(shape = "factor(MINDGDP)",
group = "factor(MINDGDP)",
colour = paste0("factor(PSIZE)")),
position = position_dodge(width = 0.25))
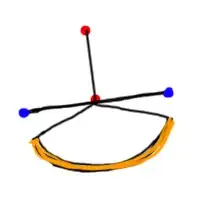
However, I want factor(METHOD_RED) on the y-axis, and auc on the x-axis. Consequently, in the following code, I have interchanged x and y, and replaced width in position_dodge with height.
ggplot2::ggplot(data = simres_auc2,
aes_string(y = "factor(METHOD_RED)",
x = "auc")) +
ggplot2::geom_point(aes_string(shape = "factor(MINDGDP)",
group = "factor(MINDGDP)",
colour = paste0("factor(PSIZE)")),
position = position_dodge(height = 0.25))
However, this code gives the following plot, in which position_dodge is not working as I had hoped.
Does anyone know why this is the case, and how I can circumvent the issue? Please note that using coord_flip is not an option for me, as it adversely affects faceting that I want to use in the code. See, for example, this question and this Github issue.