Following some discussion with @Soonts I got curious about the performance of the three approaches used in the answers: one with generating random angles, one using normally distributed coordinates, and one rejecting uniformly distributed points.
Here's my attempted comparison:
import numpy as np
def sample_trig(npoints):
theta = 2*np.pi*np.random.rand(npoints)
phi = np.arccos(2*np.random.rand(npoints)-1)
x = np.cos(theta) * np.sin(phi)
y = np.sin(theta) * np.sin(phi)
z = np.cos(phi)
return np.array([x,y,z])
def sample_normals(npoints):
vec = np.random.randn(3, npoints)
vec /= np.linalg.norm(vec, axis=0)
return vec
def sample_reject(npoints):
vec = np.zeros((3,npoints))
abc = 2*np.random.rand(3,npoints)-1
norms = np.linalg.norm(abc,axis=0)
mymask = norms<=1
abc = abc[:,mymask]/norms[mymask]
k = abc.shape[1]
vec[:,0:k] = abc
while k<npoints:
abc = 2*np.random.rand(3)-1
norm = np.linalg.norm(abc)
if 1e-5 <= norm <= 1:
vec[:,k] = abc/norm
k = k+1
return vec
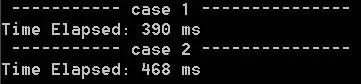
Then for 1000 points
In [449]: timeit sample_trig(1000)
1000 loops, best of 3: 236 µs per loop
In [450]: timeit sample_normals(1000)
10000 loops, best of 3: 172 µs per loop
In [451]: timeit sample_reject(1000)
100 loops, best of 3: 13.7 ms per loop
Note that in the rejection-based implementation I first generated npoints samples and threw away the bad ones, and I only used a loop to generate the rest of the points. It seemed to be the case that the direct step-by-step rejection takes a longer amount of time. I also removed the check for division-by-zero to have a cleaner comparison with the sample_normals case.
Removing vectorization from the two direct methods puts them into the same ballpark:
def sample_trig_loop(npoints):
x = np.zeros(npoints)
y = np.zeros(npoints)
z = np.zeros(npoints)
for k in range(npoints):
theta = 2*np.pi*np.random.rand()
phi = np.arccos(2*np.random.rand()-1)
x[k] = np.cos(theta) * np.sin(phi)
y[k] = np.sin(theta) * np.sin(phi)
z[k] = np.cos(phi)
return np.array([x,y,z])
def sample_normals_loop(npoints):
vec = np.zeros((3,npoints))
for k in range(npoints):
tvec = np.random.randn(3)
vec[:,k] = tvec/np.linalg.norm(tvec)
return vec
In [464]: timeit sample_trig(1000)
1000 loops, best of 3: 236 µs per loop
In [465]: timeit sample_normals(1000)
10000 loops, best of 3: 173 µs per loop
In [466]: timeit sample_reject(1000)
100 loops, best of 3: 14 ms per loop
In [467]: timeit sample_trig_loop(1000)
100 loops, best of 3: 7.92 ms per loop
In [468]: timeit sample_normals_loop(1000)
100 loops, best of 3: 10.9 ms per loop