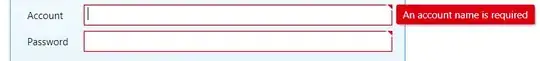
I am trying to create a plot with ggplot2 but the colorbar does not represent the actual data. The plot looks fine.
Here is my data
KEGG_Pathway Count Ratio pval_adjusted
1 Amino acid metabolism 67 11.67 1.231153e-14
2 Xenobiotics biodegradation and metabolism 31 11.07 4.492243e-06
3 Carbohydrate metabolism 54 7.78 2.940591e-05
4 Metabolism of cofactors and vitamins 34 8.76 2.439616e-04
5 Energy metabolism 23 9.58 1.488961e-03
6 Nucleotide metabolism 13 8.39 1.285896e-01
7 Metabolism of other amino acids 15 7.94 1.255625e-01
8 Biosynthesis of other secondary metabolites 20 5.17 1.000000e+00
9 Metabolism of terpenoids and polyketides 13 3.27 1.000000e+00
10 Lipid metabolism 9 2.77 1.000000e+00
And the code:
data$KEGG_Pathway <- factor(data$KEGG_Pathway, levels = rev(data$KEGG_Pathway))
myPalette <- colorRampPalette(brewer.pal(9, "BrBG"))(7)
ggplot(data, aes(Count, KEGG_Pathway)) + geom_point(aes(color=pval_adjusted, size=Ratio)) +
scale_colour_gradientn(colours = myPalette,
values = rescale(c(1.23e-14,4.49e-06,2.94e-05,2.44e-04,
1.49e-03,1.29e-01,1.26e-01,1)), limits = c(1e-14,1)) +
scale_size_area(breaks = seq(0,12, by=2)) + theme_bw()
The plot looks exactly like what I want it to. But the colorbar is completely crazy (I wanted a gradient bar showing the limits in my vector values, and all the colours in my palette, something like in here):
I have played around with guide = "colorbar" and guide_colorbar() but it produces exactly this all the time.